The aim of the LMMsolver package is to provide an
efficient and flexible system to estimate variance components using
restricted maximum likelihood or REML (Patterson and Thompson 1971), for
models where the mixed model equations are sparse. An example of an
application is using splines to model spatial (Rodríguez-Álvarez et
al. 2018; Boer, Piepho, and Williams 2020) or temporal (Bustos-Korts et
al. 2019) trends. Another example is mixed model Quantitative Trait
Locus (QTL) analysis for multiparental populations, allowing for
heterogeneous residual variance and design matrices with
Identity-By-Descent (IBD) probabilities (Li et al. 2021).
install.packages("LMMsolver")remotes::install_github("Biometris/LMMsolver", ref = "develop", dependencies = TRUE)As an example of the functionality of the package we use a model
defined in Rodríguez-Álvarez et al. (2015). It uses the
USprecip data set in the spam package (Furrer
and Sain 2010).
library(LMMsolver)
library(ggplot2)
## Get precipitation data from spam
data(USprecip, package = "spam")
## Only use observed data.
USprecip <- as.data.frame(USprecip)
USprecip <- USprecip[USprecip$infill == 1, ]A two-dimensional P-spline can be defined with the
spl2D() function, with longitude and latitude as
covariates. The number of segments chosen here is equal to the number of
segments used in Rodríguez-Álvarez et al. (2015).
obj1 <- LMMsolve(fixed = anomaly ~ 1,
spline = ~spl2D(x1 = lon, x2 = lat, nseg = c(41, 41)),
data = USprecip)The summary function gives a table with the effective dimensions and the penalty parameters:
summary(obj1)
#> Table with effective dimensions and penalties:
#>
#> Term Effective Model Nominal Ratio Penalty
#> (Intercept) 1.00 1 1 1.00 0.00
#> lin(lon, lat) 3.00 3 3 1.00 0.00
#> s(lon) 302.60 1936 1932 0.16 0.26
#> s(lat) 409.09 1936 1932 0.21 0.08
#> residual 5190.31 5906 5902 0.88 13.53
#>
#> Total Effective Dimension: 5906The spatial trend for the precipitation can now be plotted on the map of the USA.
plotDat <- obtainSmoothTrend(obj1, grid = c(200, 300), includeIntercept = TRUE)
usa = maps::map("usa", regions = "main", plot = FALSE)
v <- sp::point.in.polygon(plotDat$lon, plotDat$lat, usa$x, usa$y)
plotDat <- plotDat[v == 1, ]
ggplot(plotDat, aes(x = lon, y = lat, fill = ypred)) +
geom_tile(show.legend = TRUE) +
scale_fill_gradientn(colors = topo.colors(100))+
labs(title = "Precipitation (anomaly) US April 1948", x = "Longitude", y = "Latitude") +
coord_fixed() +
theme(panel.grid = element_blank())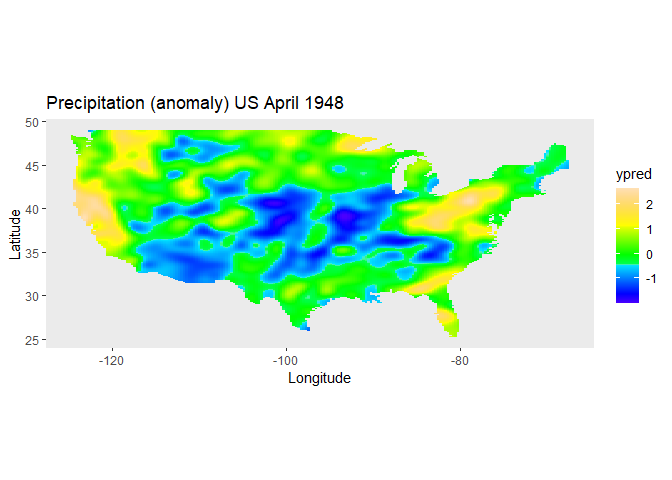
Further examples can be found in the vignette.
vignette("Solving_Linear_Mixed_Models")